Cardiomyocyte proliferation and heart regeneration
Cardiac regeneration involves the generation of new cardiomyocytes from pre-existing cardiomyocytes via proliferation. However, adult cardiomyocytes proliferate at extremely low frequency, which is not sufficient to regenerate the damaged heart, e.g. after myocardial infarction. Due to the very limited proliferation potential of cardiomyocytes, it remains technically challenging to detect their proliferation in vivo. We are developing a genetic approach for seamless recording of cardiomyocyte-specific proliferation for in situ analysis and quantification of new cardiomyocytes. We will apply the new genetic system for in vivo screening of the potential genes that regulate cell-cycle activity in adult cardiomyocytes.


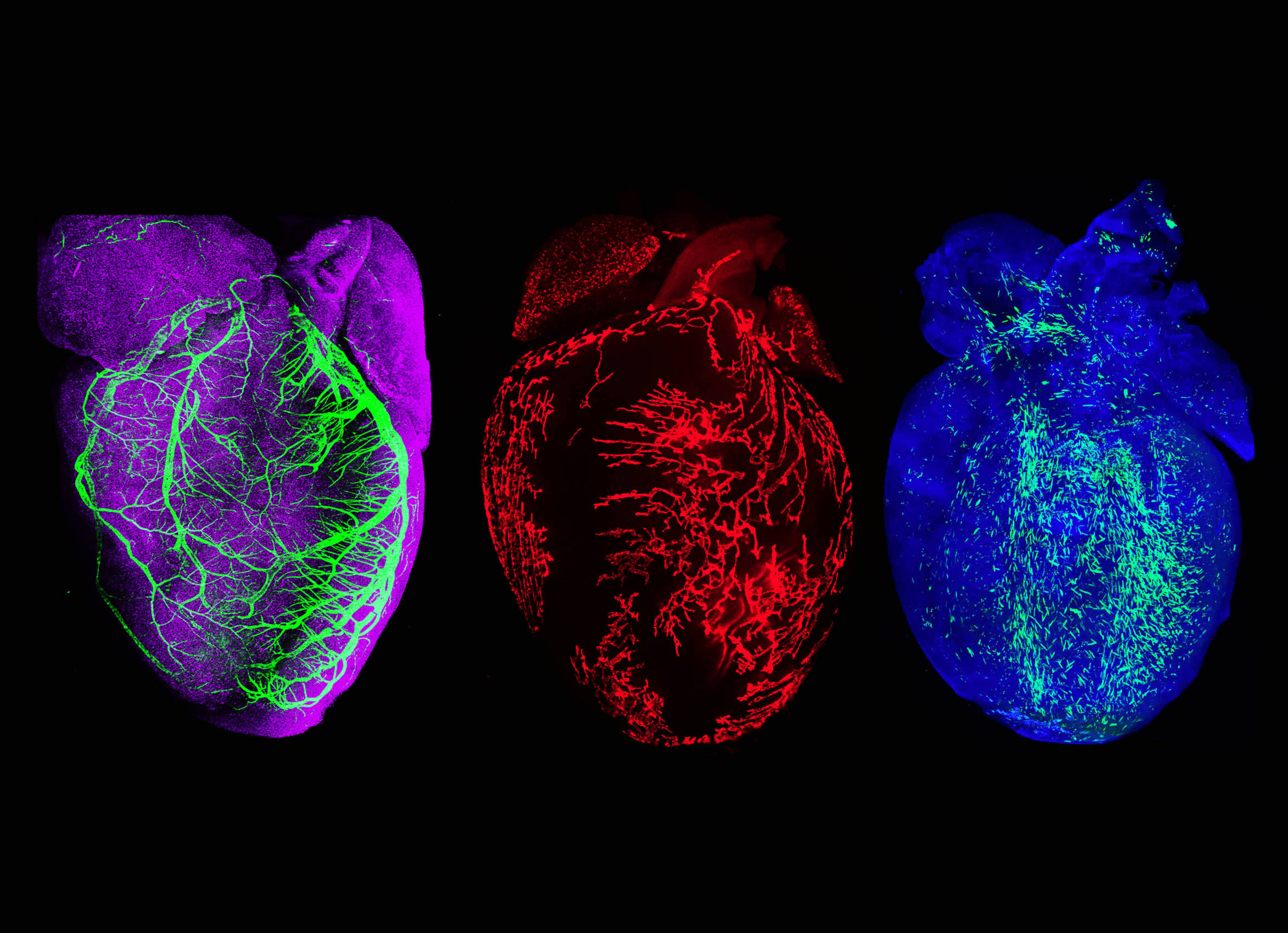
Coronary development and neovascularization
Coronary artery disease causes myocardial infarction and heart failure. Identifying coronary vascular progenitors and their developmental program inspires novel regenerative treatments for cardiac diseases. By using genetic lineage tracing, we identified two major developmental origins for coronary endothelial cells in developing hearts: sinus venosus and ventricular endocardium. In the adult heart, pre-existing coronary endothelial cells are the cellular sources for neovascularization in injured myocardium. By genetic manipulation of signaling pathways, we will elucidate the molecular mechanisms regulating coronary development and regeneration. We aim to apply this knowledge to promote coronary angiogenesis and arteriogenesis after myocardial infarction.
Genetic lineage tracing technology
Cre-loxP recombination has been widely used for in vivo cell lineage tracing. However, Cre expression is not always specific, leading to cell fate controversies in many studies. We developed a dual recombinases-mediated genetic approach by combining two orthogonal recombinases, such as Cre and Dre. Dural recombinases significantly enhance the precision of genetic lineage tracing, resolving some controversies over cell fate plasticity. We also generate dual recombinases-mediated proliferation tracing technology. Fate mapping studies using dual recombinases lead to many interesting discoveries and provide new insights on the cell fate and function of unique cell populations. We use this new technology to further explore and understand stem cell fate plasticity in tissue repair and regeneration.


Stem cells and tissue regeneration
We are interested to understand the stem cell fate plasticity and its function in tissue repair and regeneration. We have applied our newly developed genetic lineage tracing technology in exploring stem cells in other organs such as liver and lung. Recently, we used dual recombinases mediated genetic lineage tracing to specifically label stem cells that reside at the bronchioalveolar-duct junction. Fate mapping and clonal analysis showed that these bronchioalveolar stem cells became activated and responded distinctly to different lung injuries, and differentiated into multiple epithelial cell lineages for lung regeneration. We also used proliferation tracing system and found sources for new hepatocyte generation in liver homeostasis and regeneration. We aim to use new technology to understand better the stem cell biology and regenerative medicine.